
The actin cytoskeleton is essential to cellular function and hence to life. It is regulated not only via a complex synergistic and competitive interplay between actin-binding proteins (ABP), but also by filament biochemistry and filament geometry. Both the biochemical identity of an actin filament as well as the geometrical identity of actin networks are defined by ABP activity, while biochemistry and geometry also regulate ABP recruitment. However, it is still unknown how actin filament biochemistry, geometry and ABP activity jointly define actin cytoskeleton remodelling in a dynamic system such as cell migration. Our lab tries to resolve this critical relationship using cellular structural biology approaches to enable a better understanding of the multitude of cellular processes that ultimately depend on actin.
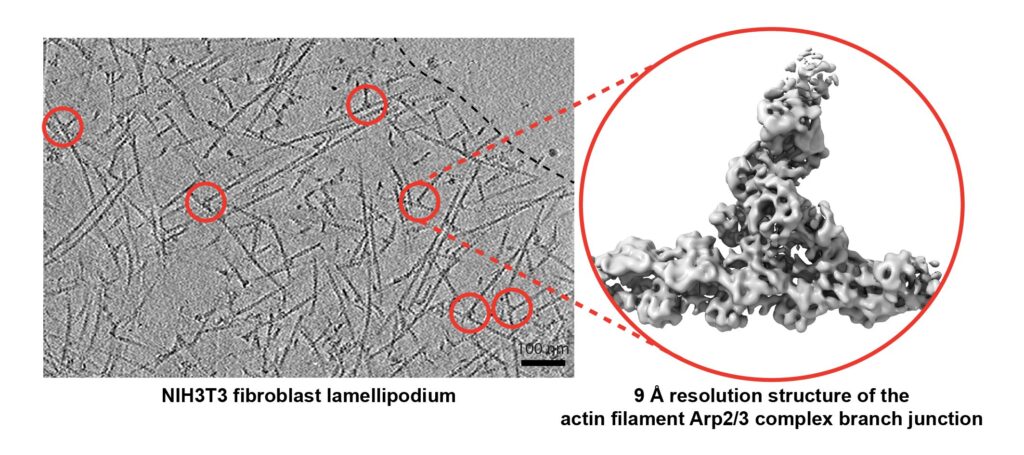
Our work lab has provided the first high-resolution in situ structure of the Arp2/3 complex branch junction (Fäßler et al., 2020a), as well as contributed to method development for cryo-ET specimen preparation (Fäßler et al., 2020b) and data analysis (Dimchev et al., 2021). Our approach performed in (Fäßler et al., 2023) to study Arp2/3 complex isoform specific regulation of cell migration presents our strategy for integrated cellular structural biology, holistically describing the role of ABPs in the actin cytoskeleton from the cell biology level to the ultrastructural level and down to the molecular layer.
ActinID – A molecular atlas of Actin filament IDentities in the cell motility machinery

In our ERC-funded project ActinID we will advance in situ structural biology to generate a molecular atlas of the actin cytoskeleton proteome in migratory cell protrusions. Our overarching aims are to:
1) Develop: cryo-electron tomography workflows to achieve 3D imaging of entire actin networks in cellular protrusions at single-filament resolution, combined with detailed quantitative analysis. This will provide the ground truth on how dynamic filament geometries steer directional cell movement.
2) Solve: high-resolution in situ structures of ABPs and F-actin and describe ABP quantity, ABP distribution and spatial correlation with their potential partners. This visual proteome will reveal how actin filament identities are regulated in an entire system.
3) Study: the reciprocal regulation of ABP activity, filament geometry and biochemistry via (genetic) manipulation of ABPs using integrative cell and structural biology experiments, and relate this to cell migration characteristics.
ActinID will be transformative for our understanding of actin cytoskeleton regulation, while also advancing the potential of in situ structural biology to go beyond isolated structural descriptions of biological systems.
cryoSPARTAN – in situ structural biology of the Actin cytoskeleton

In this FWF-funded ESPRIT project, Jesse Hansen develops novel data acquisition and image processing workflows to integrate different cryo-EM modalities to obtain high-resolution in situ structures of actin-binding proteins.